Research
Current Research Projects
Genome Evolution
 After billions of years of evolution, life on Earth has given rise to “endless forms most beautiful and most
wonderful,” as Darwin put it. My research seeks to understand how this stunning biological diversity is reflected
in, shaped by, and influences genomic information. With the advent of high-throughput sequencing and computational
biology, we can now explore genome evolution at unprecedented scales, moving beyond single-species studies to
comparative and population-level approaches that span the entire tree of life.
After billions of years of evolution, life on Earth has given rise to “endless forms most beautiful and most
wonderful,” as Darwin put it. My research seeks to understand how this stunning biological diversity is reflected
in, shaped by, and influences genomic information. With the advent of high-throughput sequencing and computational
biology, we can now explore genome evolution at unprecedented scales, moving beyond single-species studies to
comparative and population-level approaches that span the entire tree of life.
A central theme in my work is the genomic dynamics of close biological partnerships. I am particularly interested in the evolutionary forces that shape co-evolving genomes in physically intimate systems, such as those between hosts and symbionts, with a special focus on fungal and photosynthetic partners in lichens, as well as interactions between nuclear and organellar genomes within a single cell. My research spans environments ranging from tropical forests to polar regions, utilizing large-scale bioinformatic approaches to investigate how genomes adapt to ecological changes and evolve in response to long-term symbiotic associations.
1. Biodiversity Genomics: Large-Scale Discovery
 As co-director of the Grainger Bioinformatics Center at the Field Museum, I lead initiatives that use genome-scale
data to accelerate our understanding of global biodiversity. Our work spans a wide range of organisms, including
ferns, flies, weasels, rats, butterflies, and lichenized fungi. It involves sequencing hundreds of genomes and
processing thousands of museum specimens each year. This integrative approach supports questions about species
limits, trait evolution, conservation genomics, and the deep history of life.
As co-director of the Grainger Bioinformatics Center at the Field Museum, I lead initiatives that use genome-scale
data to accelerate our understanding of global biodiversity. Our work spans a wide range of organisms, including
ferns, flies, weasels, rats, butterflies, and lichenized fungi. It involves sequencing hundreds of genomes and
processing thousands of museum specimens each year. This integrative approach supports questions about species
limits, trait evolution, conservation genomics, and the deep history of life.
The Next Generation of Museomics
We apply methods such as RAD-seq, target capture, and whole-genome sequencing to study systematics, species delimitation, and evolutionary history across diverse taxa. Recent work includes uncovering cryptic diversity in polar Usnea lichens (Otero et al. 2023), revising the taxonomy of the weasel genus Neogale through mitogenomics (Patterson et al. 2025), and using ancient DNA from a 93-year-old museum specimen to confirm that the extinct Xerces blue butterfly was a distinct species (Grewe et al. 2021). That study exemplifies the power of bioinformatics in natural history collections and the importance of continued collecting for future questions and methods, as this butterfly was collected when little was known about DNA and its implications for research.
UnFATE: A Universal Toolkit for Fungal Phylogenomics and Species Delimitation
 To support biodiversity genomics efforts, we also develop software. Most recently, we created a universal probe set
and the UnFATE pipeline for phylogenomic studies of filamentous ascomycetes (Ametrano et al. 2025).
UnFATE enables large-scale phylogenomic analyses across the entire Pezizomycotina clade. The pipeline simplifies the
path from raw reads to phylogenetic trees, making it accessible even to researchers with limited bioinformatics
training. In addition to tree inference, UnFATE also supports species delimitation using the captured genes,
offering a comprehensive framework for studying fungal diversity with high resolution.
To support biodiversity genomics efforts, we also develop software. Most recently, we created a universal probe set
and the UnFATE pipeline for phylogenomic studies of filamentous ascomycetes (Ametrano et al. 2025).
UnFATE enables large-scale phylogenomic analyses across the entire Pezizomycotina clade. The pipeline simplifies the
path from raw reads to phylogenetic trees, making it accessible even to researchers with limited bioinformatics
training. In addition to tree inference, UnFATE also supports species delimitation using the captured genes,
offering a comprehensive framework for studying fungal diversity with high resolution.
2. Genome Co-evolution in Symbiotic Systems
 Lichens remain a core focus in my research as models for studying symbiosis, coevolution, and evolutionary
innovation. Using genome and transcriptome sequencing, we examine how lichen fungi and their algal partners adapt to
extreme environments and evolve reproductive strategies. Recent work uncovered unexpected genomic potential for sex
in the seemingly asexual genus Lepraria, suggesting a cryptic or parasexual reproductive mode (Doellman et al. 2024). In Usnea, we revisited the species-pair concept through population
genomics, demonstrating that reproductive mode shapes lineage divergence along a speciation continuum (Otero et al. 2025). Building on these studies, we are now investigating the genomic mechanisms and
evolutionary diversity of sexual systems in lichens.
Lichens remain a core focus in my research as models for studying symbiosis, coevolution, and evolutionary
innovation. Using genome and transcriptome sequencing, we examine how lichen fungi and their algal partners adapt to
extreme environments and evolve reproductive strategies. Recent work uncovered unexpected genomic potential for sex
in the seemingly asexual genus Lepraria, suggesting a cryptic or parasexual reproductive mode (Doellman et al. 2024). In Usnea, we revisited the species-pair concept through population
genomics, demonstrating that reproductive mode shapes lineage divergence along a speciation continuum (Otero et al. 2025). Building on these studies, we are now investigating the genomic mechanisms and
evolutionary diversity of sexual systems in lichens.
3. Plant Mitochondrial Genome Evolution
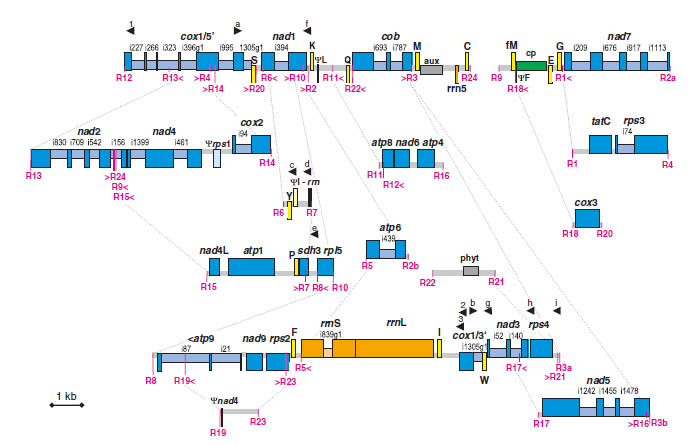 My early work focused on the extraordinary structural variation of plant mitochondrial genomes, ranging from massive
recombination in lycophytes (Grewe et al. 2009; Grewe et al. 2011; Hecht et al. 2011) to frequent genome shifts in
Brassicales (Grewe et al. 2014). These studies revealed how mitochondrial introns, splicing
mechanisms, and nuclear-mitochondrial interactions shape genome architecture. In Pelargonium, we discovered a
rare transfer of the matR gene from the mitochondrion to the nuclear genome (Grewe et al.
2016), providing a unique opportunity to study mitochondrial splicing genes using knockout techniques. We
are initiating a new project to compare mitochondrial genome evolution across lichen-forming fungi.
My early work focused on the extraordinary structural variation of plant mitochondrial genomes, ranging from massive
recombination in lycophytes (Grewe et al. 2009; Grewe et al. 2011; Hecht et al. 2011) to frequent genome shifts in
Brassicales (Grewe et al. 2014). These studies revealed how mitochondrial introns, splicing
mechanisms, and nuclear-mitochondrial interactions shape genome architecture. In Pelargonium, we discovered a
rare transfer of the matR gene from the mitochondrion to the nuclear genome (Grewe et al.
2016), providing a unique opportunity to study mitochondrial splicing genes using knockout techniques. We
are initiating a new project to compare mitochondrial genome evolution across lichen-forming fungi.
4. New Frontiers in Bioinformatics and Museum Science
Beyond traditional research questions, we use bioinformatics as a bridge between collections, public science, and emerging technologies. Projects range from urban evolutionary studies in Chicago’s brown rats (in review), to AI-assisted digitization of museum labels (Herbst et al. 2025), to training in applied genomics for conservation and education. Together, these initiatives show how genomic science can expand the research potential of museum collections and make them more relevant and accessible to diverse communities.